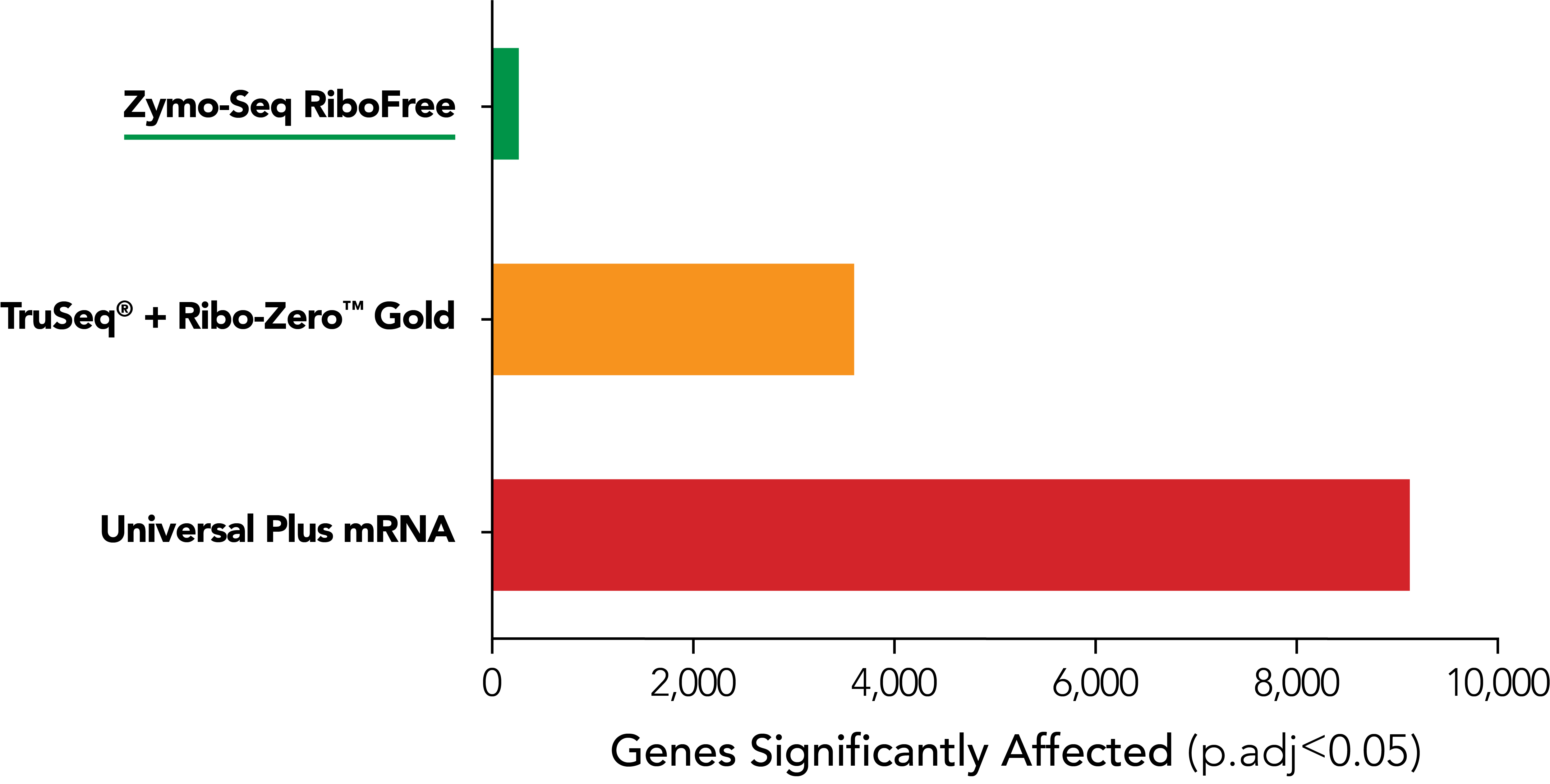
|
Sample To Sequencer In A Single Day
|
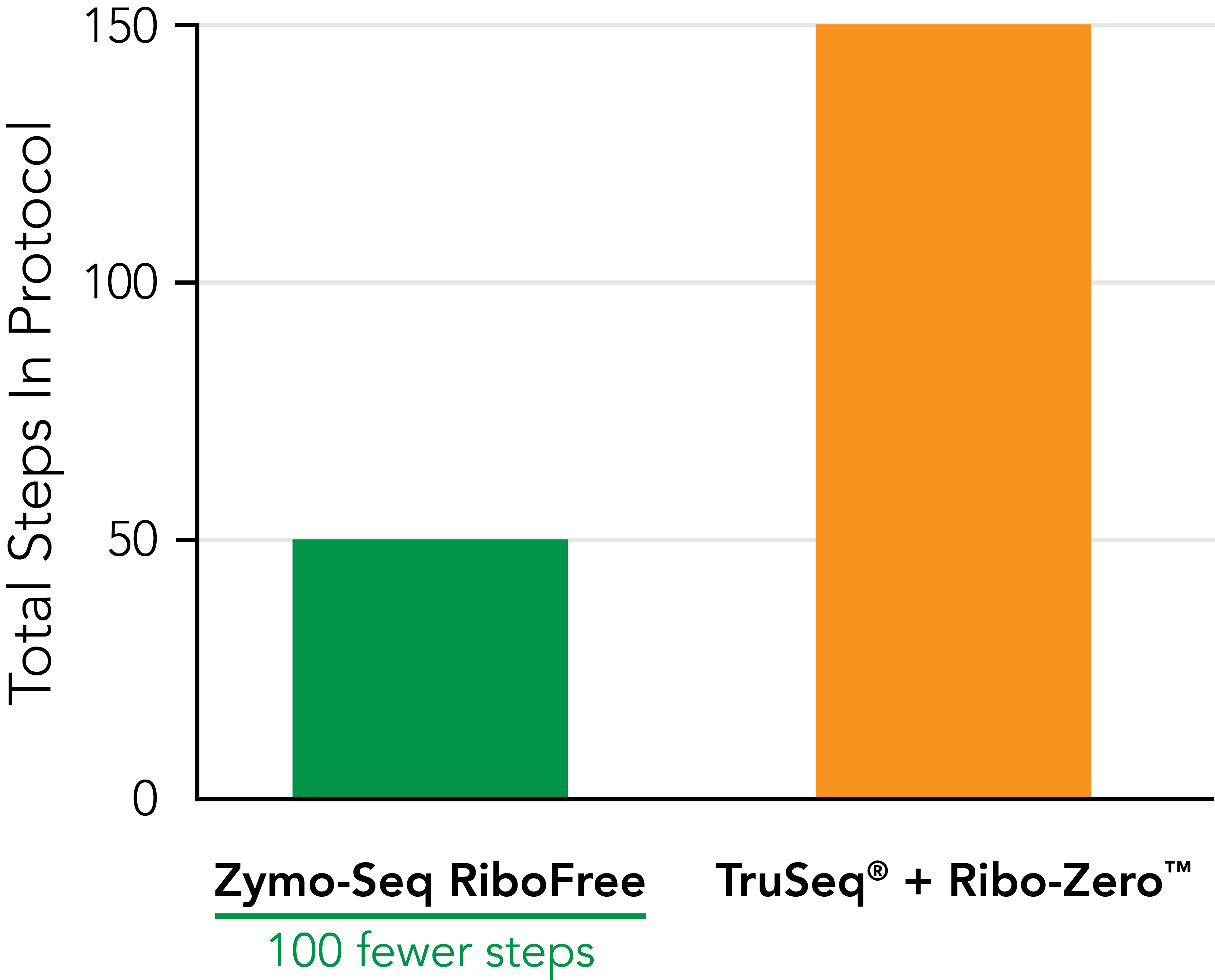
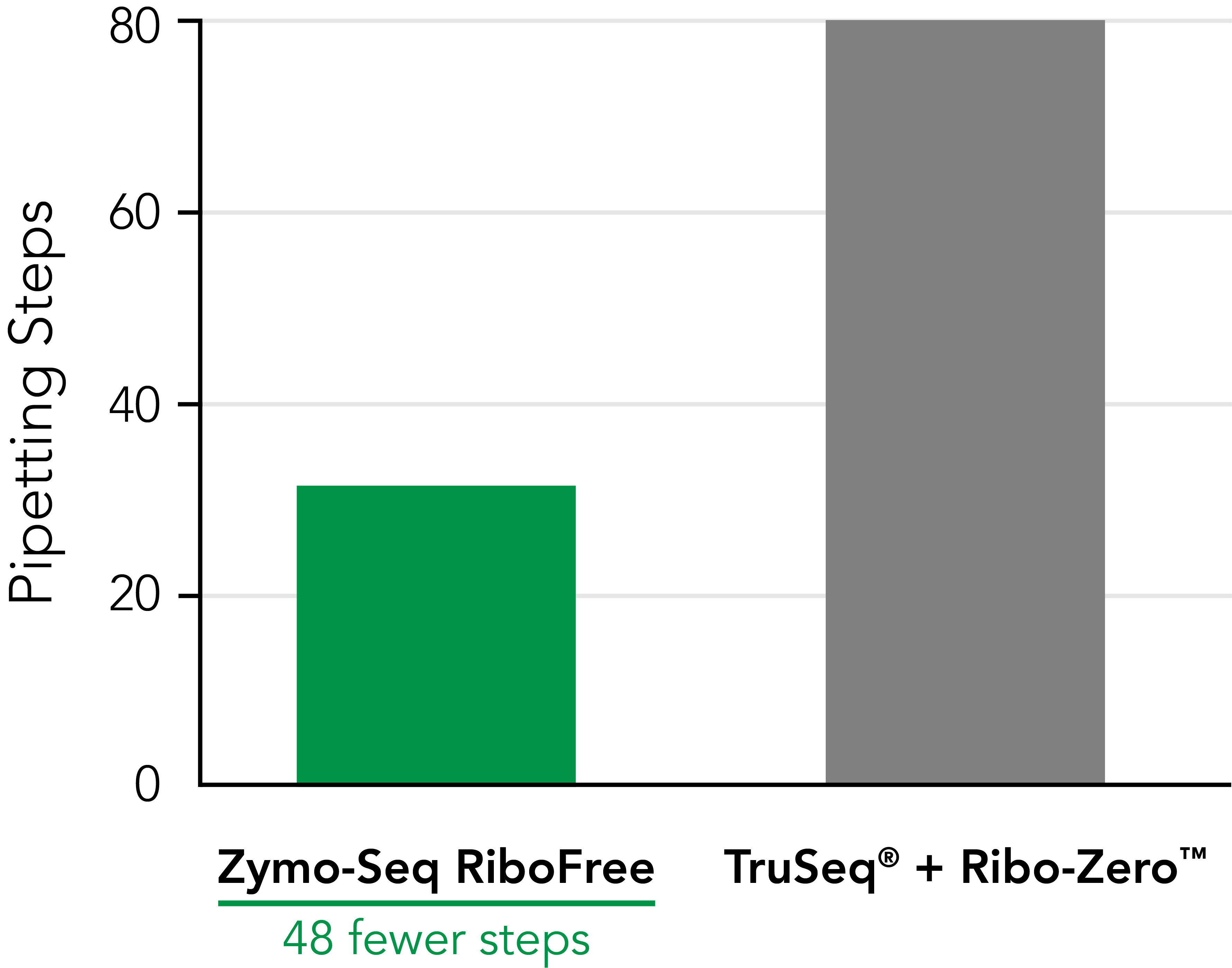
Significantly Reduce Hands‑on Time
Zymo-Seq RiboFree Total RNA Library Kit significantly reduces pipetting steps compared to TruSeq® Ribo-ZeroTM Gold. An easy-to-follow protocol allows users to go from RNA samples to the sequencer in a single day. |
Probe-free rRNA Depletion
Compatible With Any Organism
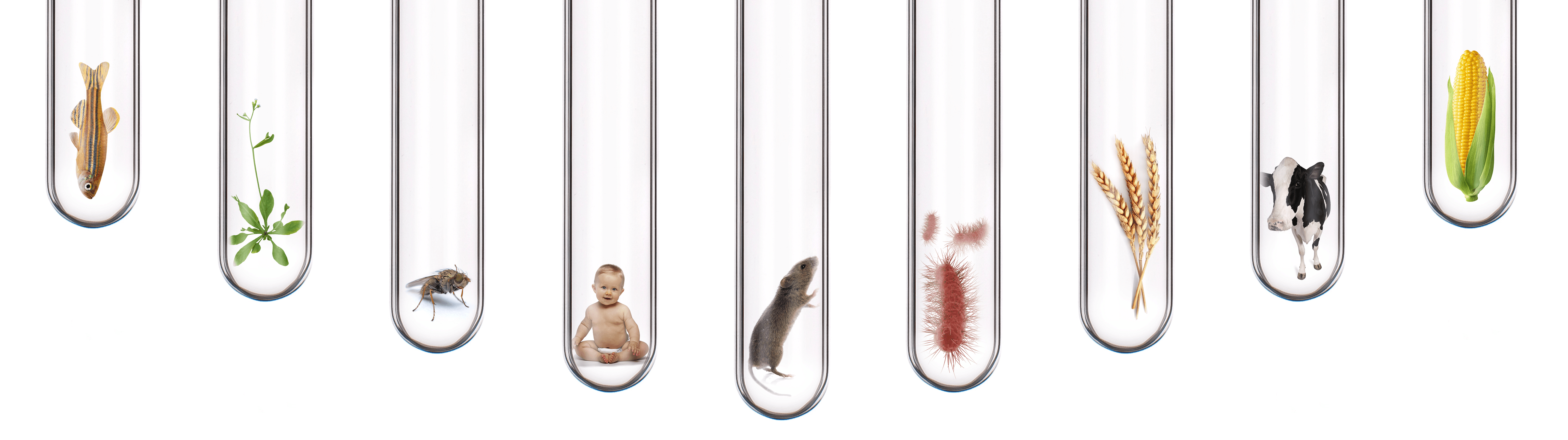
Use One Kit For Any Sample Type
Make Any Library RiboFree
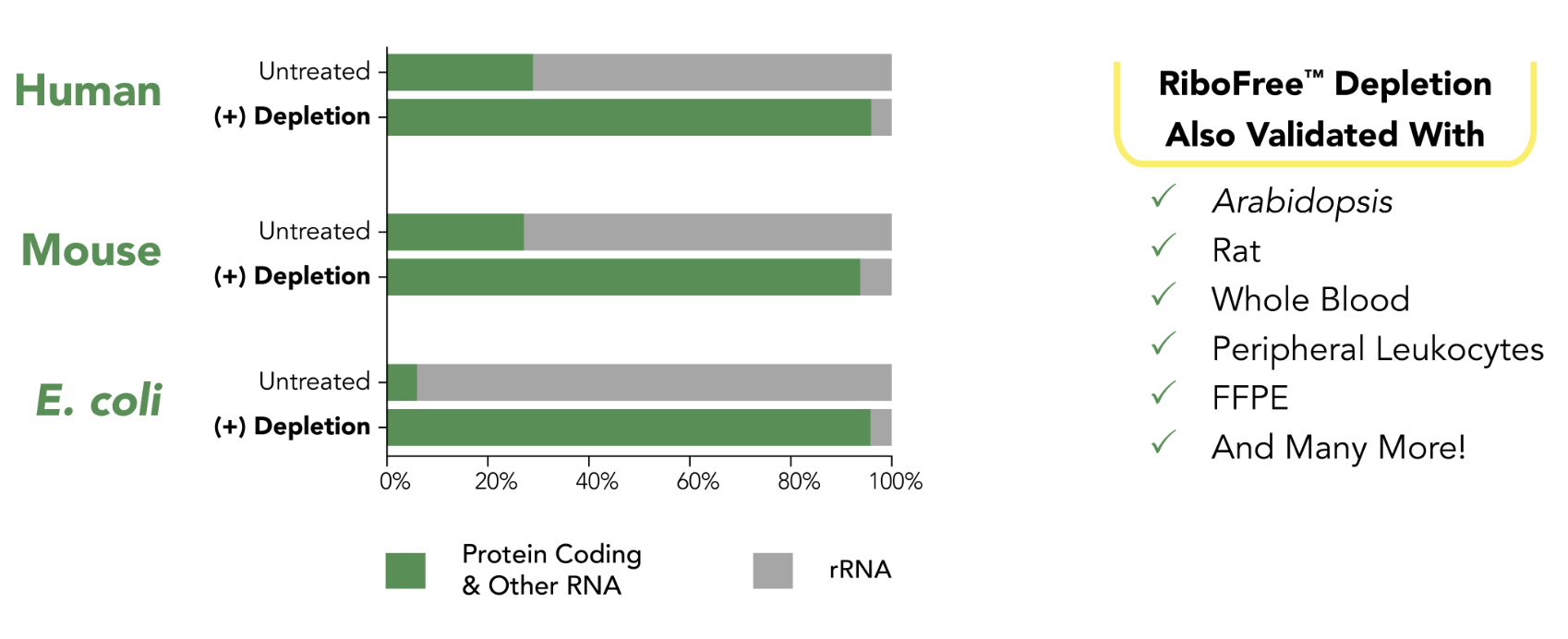
RiboFree depletion will remove rRNA from any sample type. Paired-end sequencing was performed on stranded total RNA-Seq libraries containing ERCC Spike-In Mix 1 (Life Technologies), both with and without RiboFree depletion. Read pairs were aligned to their respective genomes using the STAR aligner. Read classes were defined using a combination of Ensembl GTF gene biotypes and RepBase repeat masker annotations. Number of reads overlapping each annotation class were divided by total reads in that library to calculate percent reads of each annotation class.
Probe-free. Bias‑free. RiboFree.
The Most Accurate rRNA Depletion
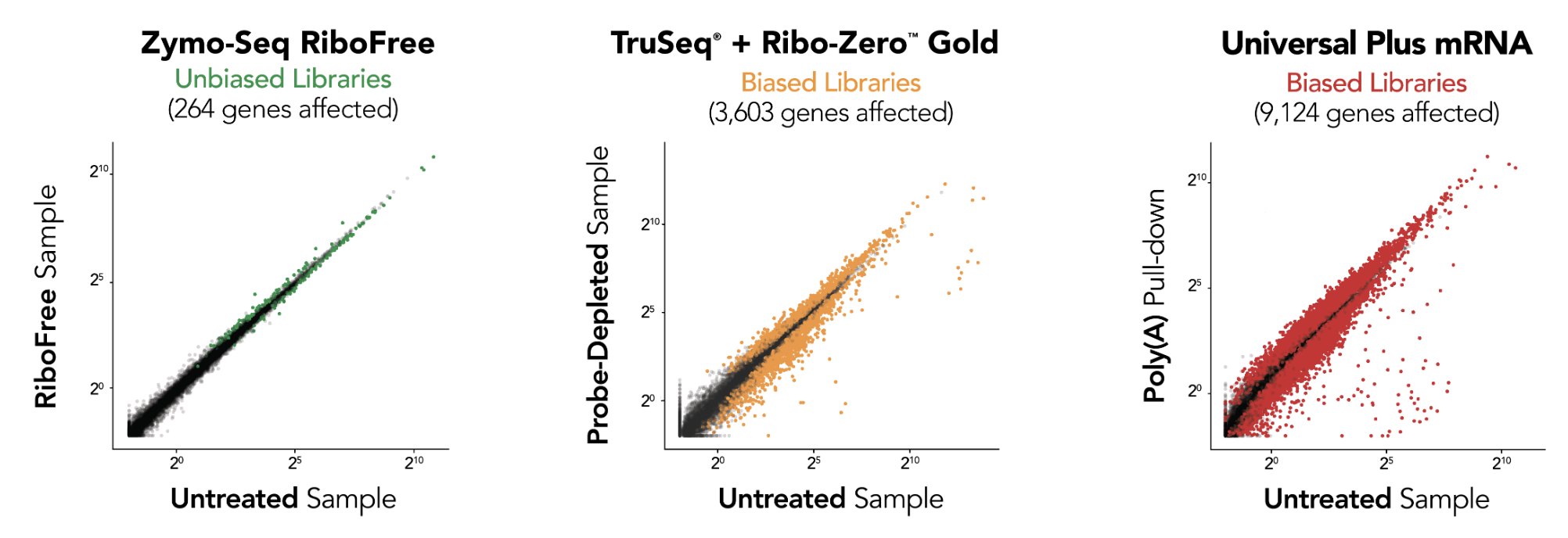
RiboFree depletion maintains native expression profiles unlike Supplier i [probe-based Ribo-ZeroTM Gold] and Supplier NG [poly(A) enrichment]. Paired-end sequencing was performed on libraries prepared from Universal Human Reference RNA (Invitrogen) containing ERCC Spike-In Mix 1 (Life Technologies), both with and without rRNA removal. Libraries were sequenced to a depth of ∼35 million reads per library, and read pairs were aligned to the hg38 human genome using the STAR aligner. The DESeq2 package was used to apply “apeglm” as a log-fold-change shrinkage correlation and to determine which of the 20,004 protein coding genes and ERCC Spike-In transcripts were significantly affected (p.adj < 0.05) by rRNA removal. Significantly affected transcripts are represented as colored dots in the scatterplots.
| Zymo Research | Supplier I | Supplier NG | |
|---|---|---|---|
| Stranded | |||
| Total time (including depletion) | 3.5 hours | >9.5 hours | >8.5 hours |
| Total steps in protocol | 51 | 151 | 127 |
| One kit. Any organism. | |||
| All reagents included from input to indexing |